Author's Note: A detailed guide on how to use Nano Banana Pro to generate scientific mechanism diagrams, pathway maps, experimental flowcharts, and other academic illustrations. Includes full prompt templates and API call code. It costs only $0.05/image via the APIYI platform.
Creating an SCI paper illustration used to take days—either hunting for a designer or manually fiddling with BioRender. Now, Nano Banana Pro can generate scientific mechanism diagrams with precise annotations directly from text descriptions. Its text rendering accuracy hits 97%+, meaning protein names, pathway labels, and bilingual annotations are all spot on, with no more garbled text.
Core Value: By the end of this post, you'll have full prompt templates for 5 common scientific drawing scenarios, plus the complete code to batch-generate images via the Nano Banana Pro API on APIYI for just $0.05 each.
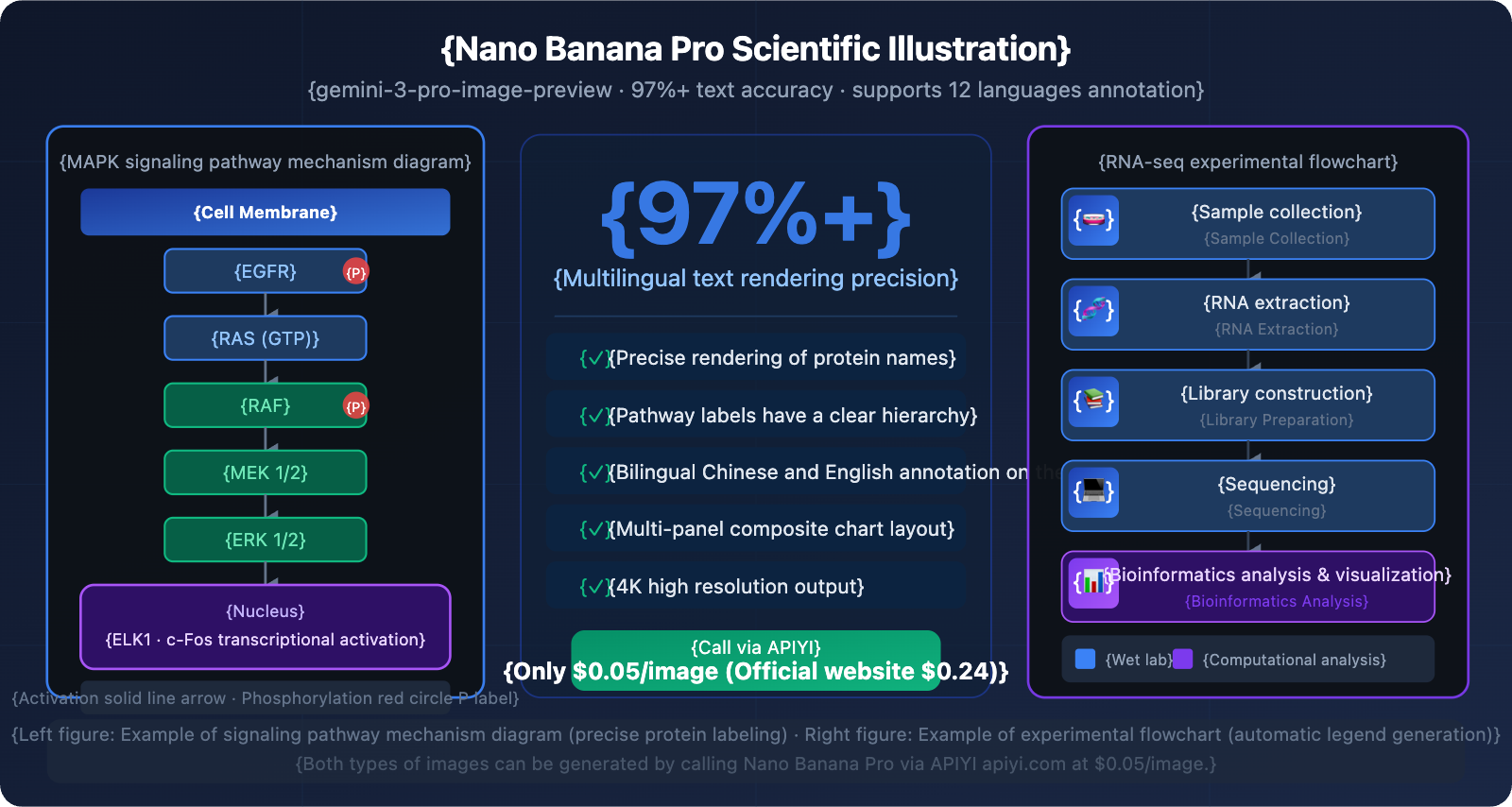
Nano Banana Pro Core Capabilities in Scientific Illustration
| Capability Dimension | Performance | Scientific Value |
|---|---|---|
| Multi-language Text Rendering | 97%+ accuracy, 12 languages | Protein names and pathway labels are precise with no garbled text |
| Scientific Annotation Understanding | Deeply trained on academic literature and textbooks | Understands professional terms like MAPK, PI3K, etc. |
| Multi-panel Composition | Supports composite figure layouts and visual hierarchy | Directly generates Figure 1 formats for papers |
| High-resolution Output | 1K / 2K / 4K levels | Meets image quality requirements for journal submissions |
| Text Embedding in Images | Automatically generates annotation blocks and legends | Saves you the trouble of adding text in Photoshop later |
Why is Nano Banana Pro the top choice for scientific illustration?
Scientific charts demand a lot from AI image tools: they don't just need to look pretty; the text annotations have to be spot-on, professional terms need to be correct, and the information hierarchy must be crystal clear. This is exactly where Nano Banana Pro holds a decisive advantage over other tools.
Looking at similar tools: DALL-E 3 slightly edges it out in anatomical realism, but it often trips up on text when dealing with complex figures full of scientific annotations. Midjourney is great for artistic flair but falls short on annotation capabilities. Adobe Firefly is more geared toward commercial design. Nano Banana Pro is currently the only AI image model that hits a high bar for both scientific text rendering and professional knowledge understanding.
Google even launched the PaperBanana framework based on this same model—it's designed specifically to turn methodology descriptions into academic illustrations automatically. It was systematically tested on 584 samples at NeurIPS 2025, which shows just how serious they are about its role in academia.
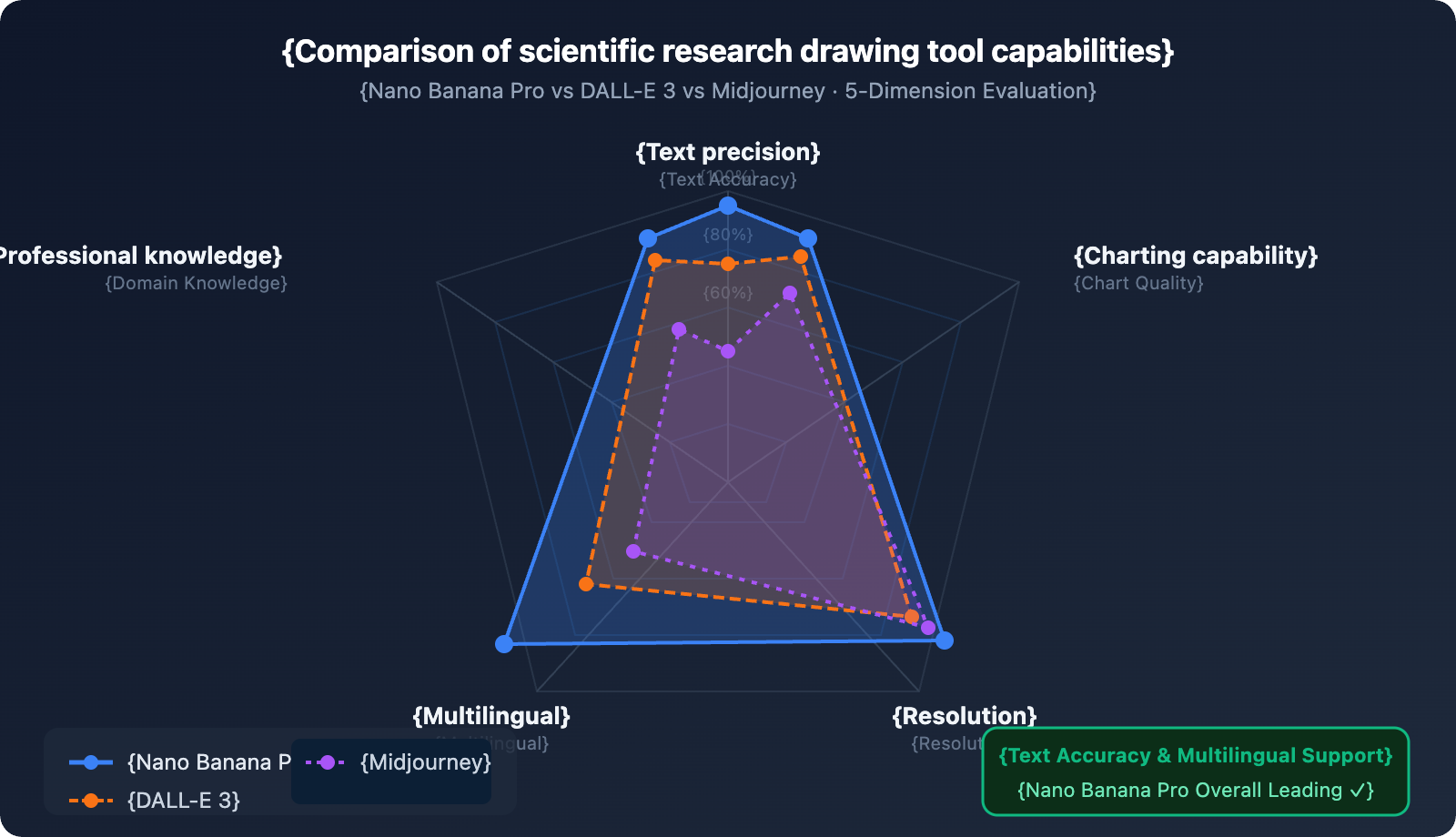
5 Scientific Drawing Scenarios and Prompt Templates
Scenario 1: Cell Signaling Pathway Mechanism
Use Cases: Cell biology and molecular biology papers; illustrating signal cascades and protein phosphorylation pathways.
Prompt Template:
Create a scientific mechanism diagram illustrating the MAPK/ERK signaling pathway.
Style: Clean white background academic illustration, suitable for SCI publication.
Layout: Vertical cascade flow from top (receptor) to bottom (nucleus).
Elements:
- Cell membrane at top with RTK receptor labeled "EGFR"
- Signal cascade: RAS → RAF → MEK → ERK with phosphorylation arrows
- Each protein shown as rounded rectangle with proper label
- Arrow styles: solid for activation, dashed for inhibition
- Nucleus at bottom with transcription factor labeled "ELK1"
Colors: Blue for membrane proteins, green for cytoplasmic kinases, red for nuclear targets
Text: All labels in English, 12pt equivalent, clearly legible
Resolution: 4K, aspect ratio 3:4
Simplified Prompt Version:
Draw a MAPK/ERK signaling pathway mechanism diagram, academic paper illustration style, white background.
Arrange vertically from top to bottom: Cell membrane (including EGFR receptor) → RAS → RAF → MEK → ERK → Nucleus (ELK1).
Represent each protein with a rounded rectangle, clearly labeled; use solid lines for activation arrows and dashed lines for inhibition.
Blue for membrane proteins, green for cytoplasmic kinases, red for nuclear transcription factors. 4K resolution, 3:4 aspect ratio.
💡 Pro-tip: For molecular pathway diagrams, it's best to explicitly specify the protein name for each node in your prompt. Nano Banana Pro renders professional terminology accurately. By calling the model via APIYI (apiyi.com), each 4K pathway diagram costs only $0.05, making it extremely cost-effective to batch-generate multiple pathway variations.
Scenario 2: Experimental Workflow
Use Cases: Methodology sections; showcasing experimental steps, sample processing flows, or instrument operation sequences.
Prompt Template:
Design a scientific experimental workflow diagram for RNA-seq analysis pipeline.
Style: Professional flowchart, dark blue background (#0f172a), white and light blue elements.
Steps (left to right, 6 steps):
1. "Sample Collection" - test tube icon
2. "RNA Extraction" - spiral RNA icon
3. "Library Preparation" - fragment icons
4. "Sequencing" - chip/sequencer icon
5. "Bioinformatics Analysis" - computer/code icon
6. "Data Visualization" - chart icon
Each step: rounded rectangle box, icon above, step name below, connected by arrows
Color coding: wet lab steps in blue, computational steps in purple
Add percentage labels for typical time allocation at each step
4K resolution, 16:9 aspect ratio, publication quality
Scenario 3: Cell Biology Structural Illustration
Use Cases: Teaching materials, review papers; illustrating organelle structures, tissue anatomy, or subcellular localization.
Prompt Template:
Create a detailed cell biology illustration showing mitochondrial structure and function.
Style: Scientific textbook illustration, clean white background.
Main elements:
- Cross-section view of mitochondrion (oval shape, 60% of frame)
- Outer membrane and inner membrane clearly labeled
- Cristae structure visible with labels
- ATP synthase complexes on inner membrane (labeled "ATP Synthase")
- Electron transport chain components (Complex I-IV) on inner membrane
- Matrix labeled with "TCA Cycle" and Krebs cycle intermediates
- Small arrows showing electron flow and proton gradient
- Legend box (bottom right): color key for different components
Text: All anatomical labels in English, bilingual caption below in Chinese and English
4K resolution, 4:3 aspect ratio
Key Prompting Technique: When describing cellular structures, using a cross-section view is much better for showing internal details. Adding a bilingual caption allows the image to be used directly for submissions to international or regional journals.
Scenario 4: Data Visualization Infographic
Use Cases: Results presentation, graphical abstracts; transforming complex data relationships into intuitive visual charts.
Prompt Template:
Create a scientific infographic summarizing CRISPR-Cas9 gene editing efficiency data.
Style: Modern scientific infographic, dark background (#1a1a2e), high contrast.
Layout: 2x2 grid of mini-charts:
Top-left: Bar chart "Editing Efficiency by Cell Type" - HEK293/iPSC/T-cell/Primary Neuron
Top-right: Pie chart "Off-target Distribution" - 3 categories with percentages
Bottom-left: Line graph "Efficiency Over Time (Days 1-14)" - two conditions
Bottom-right: Comparison table "sgRNA Design Methods" - 3 methods, 3 metrics
Color scheme: Blue (#3b82f6) for primary data, Green (#10b981) for positive results
All axis labels, titles, and data values visible and correctly spelled
Include figure title at top: "CRISPR-Cas9 Editing Performance Summary"
4K resolution, 16:9 aspect ratio
Scenario 5: Bilingual Academic Poster
Use Cases: Academic conferences, research presentations; professional posters that need to present content in both Chinese and English simultaneously.
Prompt Template:
Design a bilingual academic conference poster for a cancer immunotherapy study.
Style: Professional academic poster, dark navy background, white and gold accents.
Layout (top to bottom):
- Title bar: "Targeting PD-L1 in Triple-Negative Breast Cancer" (English)
"靶向 PD-L1 治疗三阴性乳腺癌" (Chinese, below English)
- Author line: "[Author Names] · [Institution] · [Conference Name] 2026"
- 3-column body:
Left: "Background / 研究背景" - 3 bullet points each language
Middle: Central mechanism diagram (simplified PD-1/PD-L1 pathway)
Right: "Results / 研究结果" - 2 key bar charts with bilingual labels
- Bottom: "Conclusion / 结论" banner with key finding
- QR code (bottom right) with caption "Scan for full paper"
Colors: Navy #0f172a background, gold #f59e0b accents, white text
All Chinese and English text must be correctly rendered and readable
4K resolution, 2:3 portrait aspect ratio (A0 poster format)
Quick Start: Nano Banana Pro Scientific Drawing API
Environment Setup
pip install google-generativeai pillow
Minimalist Implementation Example (Generating a Mechanism Diagram)
import google.generativeai as genai
import base64
# Call via APIYI, price is $0.05/image (official site is $0.24)
genai.configure(
api_key="YOUR_APIYI_KEY",
client_options={"api_endpoint": "vip.apiyi.com"}
)
model = genai.GenerativeModel("gemini-3-pro-image-preview")
prompt = """
Create a scientific MAPK/ERK signaling pathway diagram.
White background, academic publication style.
Show: EGFR → RAS → RAF → MEK → ERK → ELK1 cascade.
Each protein labeled clearly, arrows show activation/inhibition.
4K resolution, 3:4 aspect ratio, SCI journal quality.
"""
response = model.generate_content(
prompt,
generation_config=genai.GenerationConfig(
response_modalities=["IMAGE"],
resolution="4K",
aspect_ratio="3:4"
)
)
for part in response.candidates[0].content.parts:
if part.inline_data:
with open("mapk_pathway.png", "wb") as f:
f.write(base64.b64decode(part.inline_data.data))
print("Mechanism diagram generated: mapk_pathway.png")
🚀 Quick Start: Register at APIYI (apiyi.com) to get your API Key, then copy the code above and replace
YOUR_APIYI_KEYto run it. The platform offers free test credits, so you can generate your first scientific figure in under 5 minutes.
View Full Batch Generation Code (Includes multi-scenario, auto-naming, and error retries)
import google.generativeai as genai
import base64
import os
import time
from typing import Optional
# Initialize client - APIYI (apiyi.com), $0.05/image
genai.configure(
api_key="YOUR_APIYI_KEY",
client_options={"api_endpoint": "vip.apiyi.com"}
)
model = genai.GenerativeModel("gemini-3-pro-image-preview")
def generate_scientific_figure(
prompt: str,
output_filename: str,
resolution: str = "4K",
aspect_ratio: str = "4:3",
max_retries: int = 3
) -> Optional[str]:
"""
Generate scientific figures
Args:
prompt: Figure description (English works best)
output_filename: Output filename (.png)
resolution: 1K / 2K / 4K
aspect_ratio: 16:9 / 4:3 / 3:4 / 1:1
max_retries: Number of retries on failure
Returns:
File path on success, None on failure
"""
for attempt in range(max_retries):
try:
response = model.generate_content(
prompt,
generation_config=genai.GenerationConfig(
response_modalities=["IMAGE"],
resolution=resolution,
aspect_ratio=aspect_ratio
)
)
for part in response.candidates[0].content.parts:
if part.inline_data and part.inline_data.mime_type.startswith("image/"):
os.makedirs(os.path.dirname(output_filename) or ".", exist_ok=True)
with open(output_filename, "wb") as f:
f.write(base64.b64decode(part.inline_data.data))
return output_filename
except Exception as e:
print(f" Attempt {attempt + 1}/{max_retries} failed: {e}")
if attempt < max_retries - 1:
time.sleep(5)
return None
# Predefined scientific figure task list
FIGURE_TASKS = [
{
"name": "MAPK Signaling Pathway",
"filename": "figures/fig1_mapk_pathway.png",
"resolution": "4K",
"aspect_ratio": "3:4",
"prompt": """
Scientific diagram: MAPK/ERK signaling pathway.
White background, academic style.
Top to bottom: EGFR receptor → RAS (GTP-bound) → RAF → MEK1/2 → ERK1/2 → nucleus (ELK1, c-Fos).
Each component: rounded rectangle, bold protein name label.
Activation arrows: solid black. Phosphorylation: circled 'P' symbol in red.
Color: blue membrane, green cytoplasm proteins, red nuclear targets.
Add "MAPK/ERK Signaling Pathway" title at top.
Publication quality, 4K.
"""
},
{
"name": "RNA-seq Experimental Workflow",
"filename": "figures/fig2_rnaseq_workflow.png",
"resolution": "4K",
"aspect_ratio": "16:9",
"prompt": """
Scientific workflow diagram: RNA-seq analysis pipeline.
Dark blue background, white icons and text.
6 steps left to right with connecting arrows:
1. Sample Collection (test tube icon)
2. RNA Extraction (helix icon)
3. Library Prep (fragment icon)
4. Sequencing (chip icon)
5. Bioinformatics (terminal icon)
6. Visualization (chart icon)
Steps 1-4 in blue (wet lab), steps 5-6 in purple (computational).
Each step: icon + step name + time estimate below.
Title: "RNA-seq Analysis Workflow". 4K, 16:9.
"""
},
{
"name": "Mitochondrial Structure",
"filename": "figures/fig3_mitochondria.png",
"resolution": "4K",
"aspect_ratio": "4:3",
"prompt": """
Scientific cell biology illustration: mitochondrion cross-section.
White background, textbook illustration style.
Detailed cross-section showing: outer membrane, inner membrane, cristae,
matrix with TCA cycle label, ATP synthase complexes (labeled) on inner membrane,
electron transport chain (Complex I-IV, labeled individually).
Small curved arrows showing proton gradient and electron flow.
Legend bottom-right: color key.
Bilingual labels: English primary, Chinese translation in parentheses.
Title: "Mitochondrial Structure and Function / 线粒体结构与功能".
4K, 4:3 ratio.
"""
},
]
def batch_generate_figures(tasks: list, output_dir: str = "figures") -> dict:
"""Batch generate scientific figures"""
results = {"success": [], "failed": []}
total = len(tasks)
print(f"\nStarting batch generation of {total} scientific figures...")
print(f"Platform: APIYI (apiyi.com) | Price: $0.05/image | Estimated total cost: ${total * 0.05:.2f}\n")
for i, task in enumerate(tasks, 1):
print(f"[{i}/{total}] Generating: {task['name']}")
path = generate_scientific_figure(
prompt=task["prompt"],
output_filename=task.get("filename", f"{output_dir}/figure_{i:02d}.png"),
resolution=task.get("resolution", "4K"),
aspect_ratio=task.get("aspect_ratio", "4:3")
)
if path:
results["success"].append({"name": task["name"], "path": path})
print(f" ✓ Done: {path}")
else:
results["failed"].append(task["name"])
print(f" ✗ Failed: {task['name']}")
# Avoid rate limits
if i < total:
time.sleep(2)
print(f"\nGeneration complete: {len(results['success'])} successful, {len(results['failed'])} failed")
print(f"Actual cost: ${len(results['success']) * 0.05:.2f}")
return results
if __name__ == "__main__":
results = batch_generate_figures(FIGURE_TASKS)
💰 Cost Comparison: Generating 20 scientific figures (a complete set for a paper) costs $4.80 via Google's official channel, but only $1.00 via APIYI (apiyi.com)—a 79% saving. For research scenarios requiring frequent iterations and modifications, the cost advantage is massive.
Advanced Prompting Techniques for Scientific Drawing
The Prompt Structure Formula
A high-quality scientific prompt = Subject Definition + Style Specifications + Element Checklist + Labeling Requirements + Resolution Parameters
| Element | Example | Role |
|---|---|---|
| Subject Definition | "Scientific diagram of CRISPR-Cas9 mechanism" | Tells the model the professional field |
| Style Specifications | "White background, academic publication style" | Controls the overall visual style |
| Element Checklist | "Show: Cas9 protein, sgRNA, target DNA, PAM sequence" | Ensures key content isn't missed |
| Labeling Requirements | "All components labeled in English, bilingual Chinese caption" | Precisely controls text content |
| Resolution Parameters | "4K resolution, 16:9 aspect ratio" | Meets submission specifications |
Common Errors and Fixes
Issue 1: Misplaced text labels
❌ Bad prompt: "label the proteins"
✅ Fixed prompt: "Each protein shown as rectangle with bold label text centered inside,
all labels: Arial 12pt equivalent, black text on white background"
Issue 2: Confusing pathway directions
❌ Bad prompt: "show the signaling cascade"
✅ Fixed prompt: "Top-to-bottom vertical flow: step 1 at top → step 2 → step 3 at bottom,
directional arrows pointing downward between each component"
Issue 3: Lack of color distinction
❌ Bad prompt: "use different colors for each component"
✅ Fixed prompt: "Color coding: membrane proteins #3b82f6 (blue), cytoplasmic #10b981 (green),
nuclear #f97316 (orange), inhibitory elements #ef4444 (red)"
Recommended Resolutions and Aspect Ratios for Different Disciplines
The aspect ratio of a scientific figure directly impacts the layout. Here are the recommended parameters organized by submission scenario:
| Use Case | Recommended Aspect Ratio | Recommended Resolution | Description |
|---|---|---|---|
| Single-column journal figure | 4:3 |
4K | Single-column illustration for Nature/Science |
| Double-column journal figure | 16:9 |
4K | Wide composite figure spanning columns |
| Vertical mechanism diagram | 3:4 |
4K | Vertical diagrams like signaling pathways or trees |
| Academic conference poster | 2:3 |
4K | A0/A1 size posters (portrait) |
| PPT/Slide illustration | 16:9 |
2K | Figures for presentations |
| Square schematic | 1:1 |
2K | Social media posts or supplementary materials |
| Banner flowchart | 21:9 |
4K | Long-process experimental step diagrams |
From Prompt to Publication: A Complete Workflow for Paper Illustrations
Let's take a cell biology paper as an example of a complete illustration workflow:
Step 1: Plan the figure structure (Zero API cost)
Determine the type and number of figures needed:
- Figure 1: Research model mechanism (signaling pathway) → 3:4 Vertical
- Figure 2: Experimental design flowchart → 16:9 Horizontal
- Figure 3A-C: Multi-panel data results → 16:9, 2×3 layout
- Supplementary: Cell structure schematics × 2 → 4:3
Total of 6 figures, total cost on APIYI: $0.30
Step 2: Generate and iterate one by one ($0.05 per run)
# Generate → Check → Refine prompt → Regenerate
# Typical iterations: 2-3 times per figure, total cost per figure $0.10-$0.15
Step 3: Accuracy check (An essential step)
Cross-check against KEGG / UniProt / Reactome databases:
- Protein name spelling ✓
- Pathway direction and arrow meanings ✓
- Correctness of interaction relationships ✓
Step 4: Format processing
4K PNG → Journal-required 300 DPI TIFF (convert using GIMP or Photoshop)
If vector graphics are needed: Import PNG into Adobe Illustrator for tracing
🎯 Efficiency Comparison: Traditionally, hand-drawing a set of 6 paper illustrations takes 2-5 business days. Using Nano Banana Pro + APIYI, the entire process takes about 2-4 hours and costs less than $1. We recommend getting an API Key through APIYI (apiyi.com) and applying these templates directly to drastically shorten your paper preparation cycle.
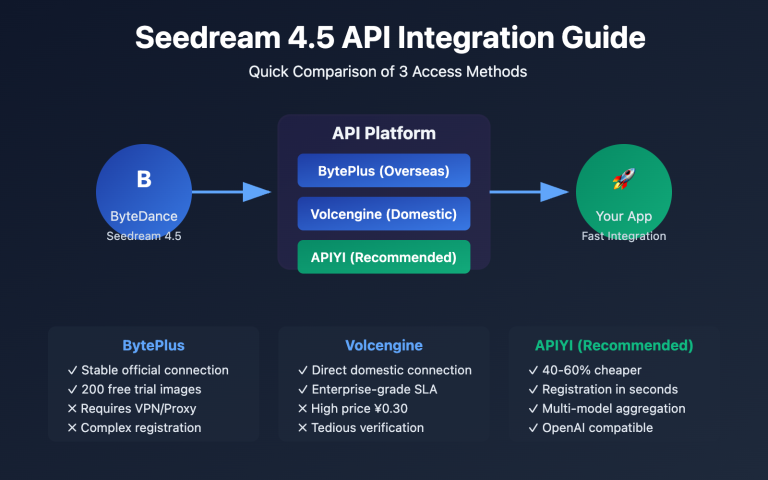
Platform Pricing and API Comparison
| Access Method | Price per 4K | Stability | Format Support | Best Use Case |
|---|---|---|---|---|
| Google Official | $0.24/req | ★★★★★ | Native Format | Enterprise / High Budget |
| APIYI | $0.05/req | ★★★★★ | Native Format ✅ | Scientific Batch Generation |
| KIE.ai | $0.12/req | ★★★☆☆ | Custom Format | Occasional Use |
| PiAPI | $0.18/req | ★★★★☆ | Custom Format | Commercial Use |
🎯 Pro Tip: For researchers and labs, paper revisions often require constant adjustments to figures, which means a high demand for batch generation. We recommend using APIYI (apiyi.com) to handle your batch image generation at the lowest possible cost. The platform supports Google's native Gemini format, so there's no need for format conversion—it's fully compatible with all the code examples in this guide.
FAQ
Q1: Can research figures generated by Nano Banana Pro be submitted directly to SCI journals?
Yes, you can use them, but keep the following points in mind:
- Accuracy checks are mandatory: AI-generated pathway diagrams might contain detailed errors (like unusual branches or fictional protein interactions). Before submitting, always verify them against authoritative databases like UniProt, KEGG, or Reactome.
- Resolution requirements: Opt for 4K output. Most journals require at least 300 DPI, and 4K images easily meet this standard.
- AI usage disclosure: Top-tier journals like Nature and Science require you to declare the use of AI tools for generating illustrations in the Methods section.
- Vector graphics requirements: Some journals require SVG/EPS vector formats. Nano Banana Pro outputs raster images, so you might need extra processing for those specific journals.
Images generated via APIYI (apiyi.com) are identical in quality to the official version and are perfectly suitable for preprints, non-top-tier journal submissions, and teaching materials.
Q2: How do I generate multi-panel composite figures (e.g., Figure 1A-1F)?
Nano Banana Pro supports generating multi-panel layouts in a single image. You just need to be very specific in your prompt:
Create a multi-panel scientific figure (2 rows × 3 columns, labeled A-F):
Panel A (top-left): [Describe content of Panel A]
Panel B (top-center): [Describe content of Panel B]
Panel C (top-right): [Describe content of Panel C]
Panel D (bottom-left): [Describe content of Panel D]
Panel E (bottom-center): [Describe content of Panel E]
Panel F (bottom-right): [Describe content of Panel F]
Each panel: white background, bold letter label top-left (A, B, C...),
thin gray border between panels, consistent color scheme.
4K resolution, 16:9 aspect ratio.
If the content of each sub-figure varies significantly, you can also generate them separately and then merge them using Python's PIL library. Batch generating multiple sub-figures via APIYI (apiyi.com) costs only about $0.30 (6 images × $0.05).
Q3: Chinese labels often appear garbled or incomplete. How can I fix this?
Nano Banana Pro has excellent Chinese rendering capabilities, but it requires the right prompt phrasing:
- Explicitly declare the language: Add
"Chinese text must be correctly rendered in Simplified Chinese characters"to your prompt. - Specify font style: Use
"Use standard Song/Hei font style for Chinese labels". - Control text density: Don't overdo the Chinese labels. Keep the important ones and move secondary information to the legend.
- Bilingual strategy: Use English as the primary label with Chinese in parentheses, e.g.,
"Mitochondria (线粒体)". Chinese rendering is often more stable when English leads the way.
If the problem persists, you can generate an English version first, then use APIYI (apiyi.com) to send an image-to-image request, asking it to "replace all English labels in this image with Chinese."
Summary
Here are the core takeaways for using Nano Banana Pro for scientific illustration:
- Text precision is the biggest advantage: With 97%+ multilingual accuracy, protein names and pathway labels are rendered precisely—outperforming almost all similar tools.
- 5 scenario-specific prompt structures: Use the "vertical cascade flow" template for signaling pathways, "horizontal step flow" for experimental workflows, and clear panel layouts for composite figures.
- Choose APIYI for pricing: At $0.05 per image (compared to $0.24 on the official site), the cost of generating figures for a paper drops by 79%. It also supports direct calls in Google's native format.
- Always check accuracy before submission: AI images can contain professional detail errors, so verify them against authoritative databases.
We recommend setting up an automated scientific illustration workflow via APIYI (apiyi.com). Save the code from this article as a template and swap out prompts as needed for batch generation—it'll significantly boost your paper preparation efficiency.
References
-
Google Gemini API Image Generation Documentation
- Link:
ai.google.dev/gemini-api/docs/image-generation - Description: Official parameter guide for Nano Banana Pro, including best practices for scientific image generation.
- Link:
-
Introduction to Google's PaperBanana Framework
- Link:
the-decoder.com/googles-paperbanana-uses-five-ai-agents-to-auto-generate-scientific-diagrams - Description: Technical details of Google's multi-agent framework for automated academic illustration generation.
- Link:
-
Nano Banana Pro Life Sciences Application Review
- Link:
intuitionlabs.ai/articles/gemini-nano-banana-pro-life-sciences - Description: System performance evaluation for cell biology and molecular biology scenarios.
- Link:
-
APIYI Platform Integration Documentation
- Link:
docs.apiyi.com - Description: Instructions for obtaining API keys, Gemini native format integration, and pricing details.
- Link:
Author: Technical Team
Technical Exchange: Feel free to share your scientific illustrations in the comments! For more AI drawing tips, visit the APIYI (apiyi.com) technical community.